

Gene analysis and manipulation
Visual Gene Developer has several predefined functions as shown in the below image and table.
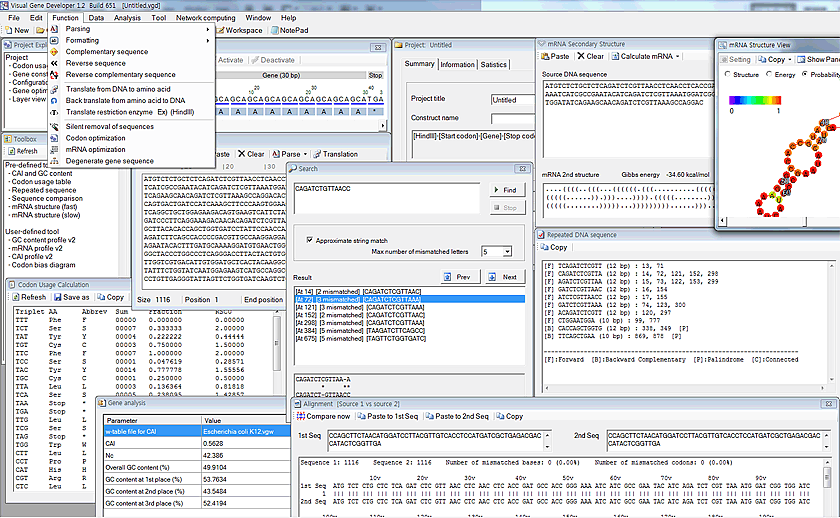
Gene analysis
| Function | Note |
| Codon usage count | |
| CAI |
Codon Adaptation index * 'Codon usage and w-table' should be defined and set to calculate CAI |
| tAI | tRNA adaptation index |
| GC content | |
| Nc |
Effective number of codons |
| Sequence comparison | Simple comparison between two sequences |
| Repeated sequence search | |
| mRNA secondary structure prediction | For more details, Click here. |
| Profiling functions: CAI, mRNA binding energy, and GC content | |
| Codon bias diagram for multiple genes | Graphical representation of codon bias based on RSCU |
| Approximate sequence search | Search similar sequences |
Sequence manipulation
| Function | Note |
|
Sequence parsing |
Remove unnecessary letters for DNA or Amino acids |
|
Formatting |
Space delimited triplet, Replace U with T, or T with U Convert to upper or lower case |
|
Translation from DNA to amino acid |
|
|
Back translation from amino acid to DNA |
|
|
Sequence reversing |
|
|
Complimentary sequence |
|
|
Silent removal of multiple sequences |
Remove multiple sequences from a test sequence in the 'Workspace' window * Silent removal means removal of certain target DNA sequence from a source sequence by replacing certain codons with synonymous codons while maintaining the expression of the same amino acids |
|
Degenerate sequence |
|
In this section, we explain about main tool menus and the class (name: GeneService) for module development.
o Menu function
All functions are in the 'Analysis' of the main menu. Before clicking on one of the functions, first, paste any sequence to the 'Workspace' window.
Codon Usage Calculation
1. First, paste sequence to the 'Workspace' window
2. Click on the 'Codon Usage Table' function.
3. You can see the result in the 'Codon Usage Calculation' window
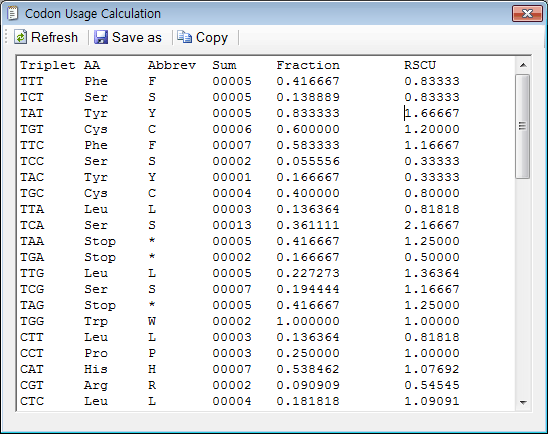
CAI, GC content and Nc
1. In order to calculate CAI, 'Codon usage and w-table' must be defined and set. First, click on the 'Project' item in the 'Project Explorer' window.
2. As an example, you can choose one of the predefined ones in the 'Project' window and then click on the 'Apply' or 'OK' button.
3. Click on the 'CAI and GC content' in the 'Analysis' menu.
4. The resulting window will be shown. Don't forget to paste the sequence to the 'Workspace' window.
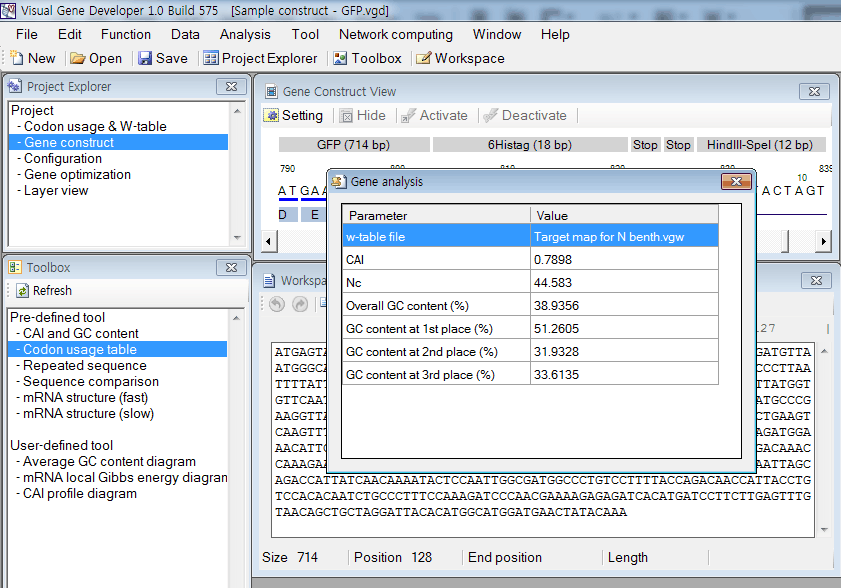
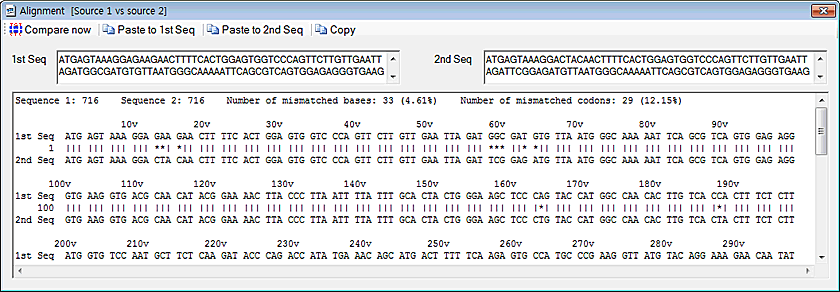
Sequence comparison
1. Click on the 'Sequence comparison' in the 'Analysis' menu.
2. You can also click on the 'Paste to 1st Seq' or 'Paste to 2nd Seq' button to paste sequence therefrom the clipboard
3. Click on the 'Compare now' button. You can see the result.
tAI (tRNA adaptation index) calculation
1. Paste any sequence to the 'Workspace' window
2. Click on 'tRNA adaptation index (tAI) in the menu
3. You can see the result in the 'tRNA adaptation index (tAI) window
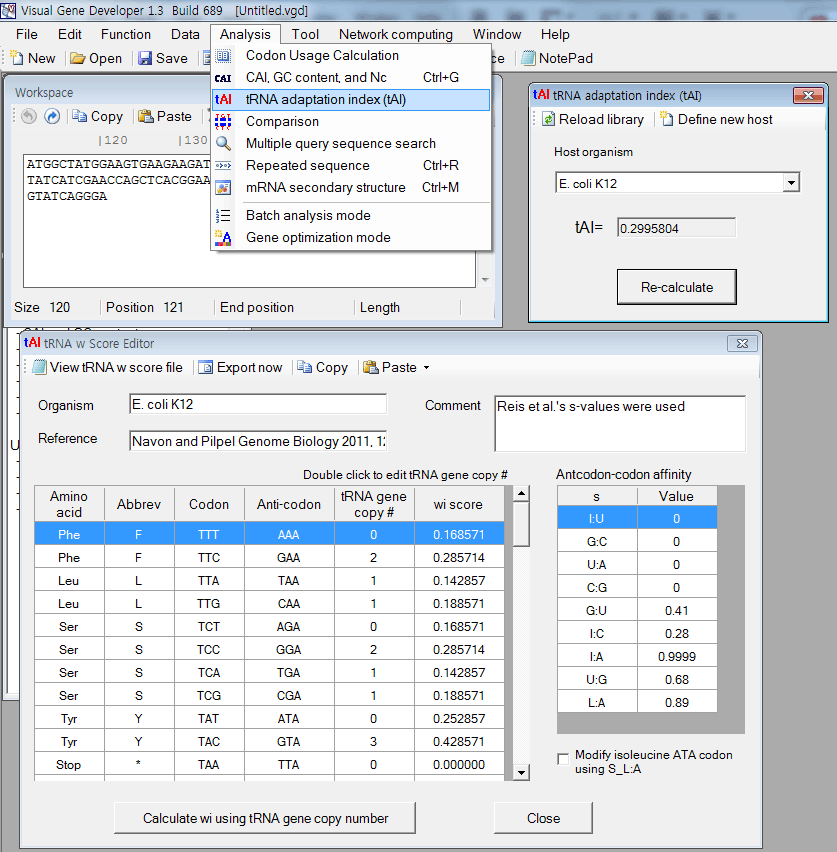
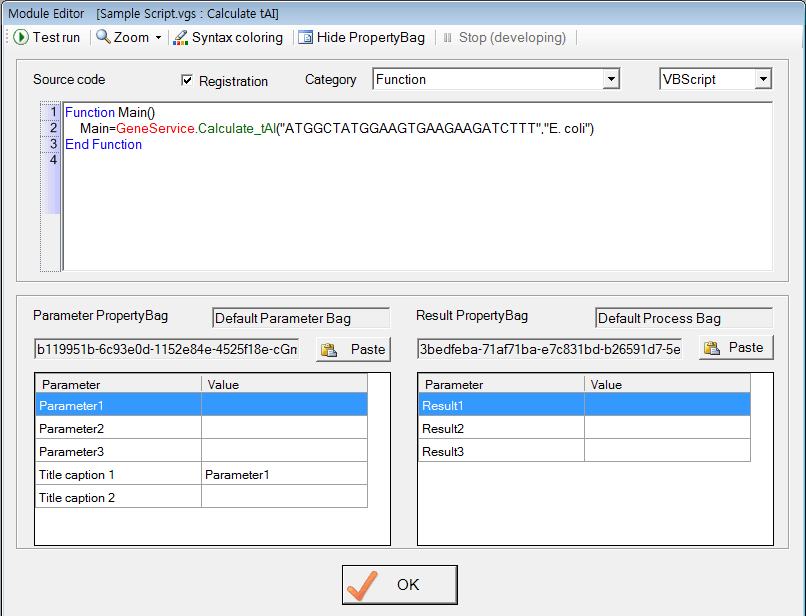
Optionally, you can call the function while doing programming in the 'Module Editor' window
Approximate sequence search
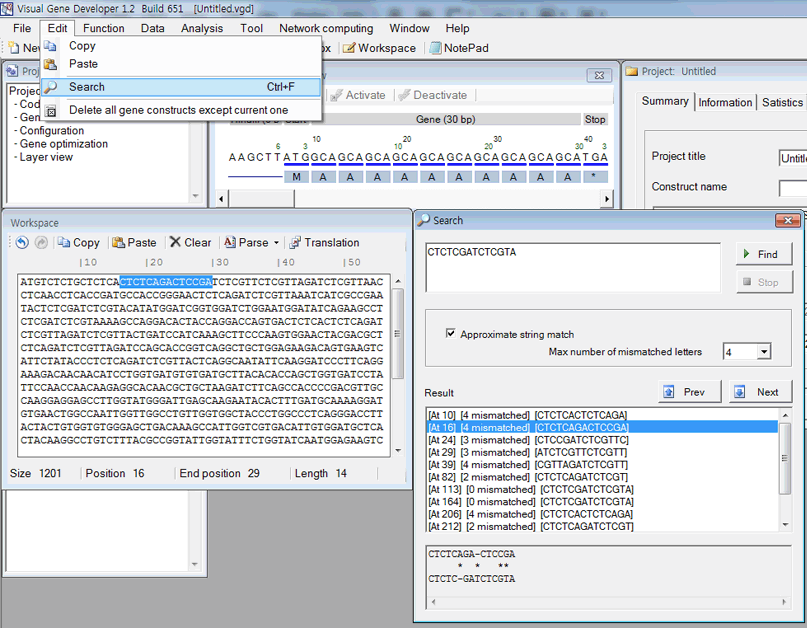
Multiple query sequence search
1. First, paste sequence to the textbox in the 'Workspace' window.
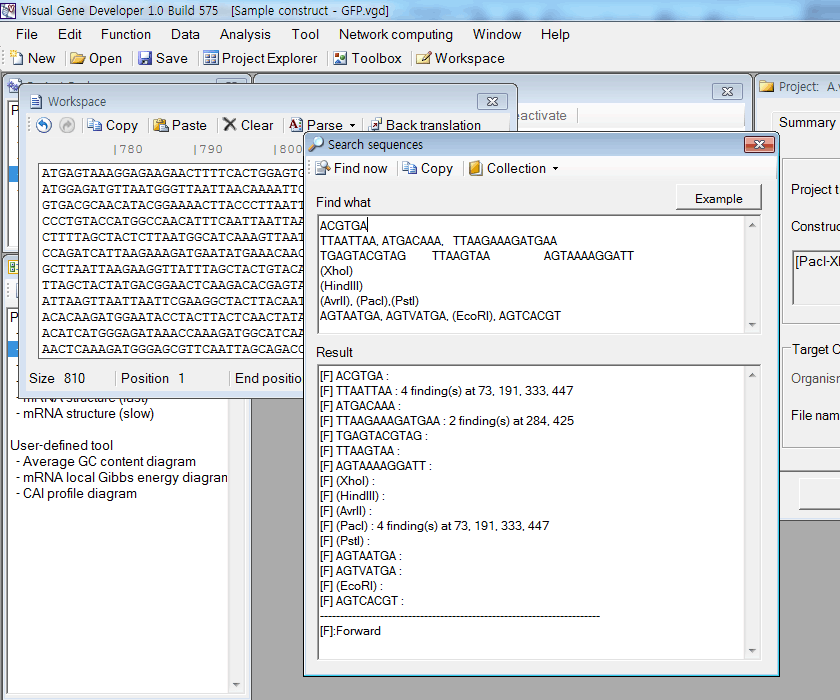
2. Click on the 'Search sequences' in the 'Analysis' menu.
3. Input any sequences to the 'FindWhat' section.
4. Click on the 'Find now' button.
Repeated sequence search
1. First input sequence to the textbox in the 'Workspace' window.
2. Click on the 'Repeated sequence' in the 'Analysis' menu. The result will be shown in the 'Repeated DNA sequence' window.
3. You can click on the 'Analyze' button in the 'Repeated DNA sequence' window instead of clicking the function in the main menu.
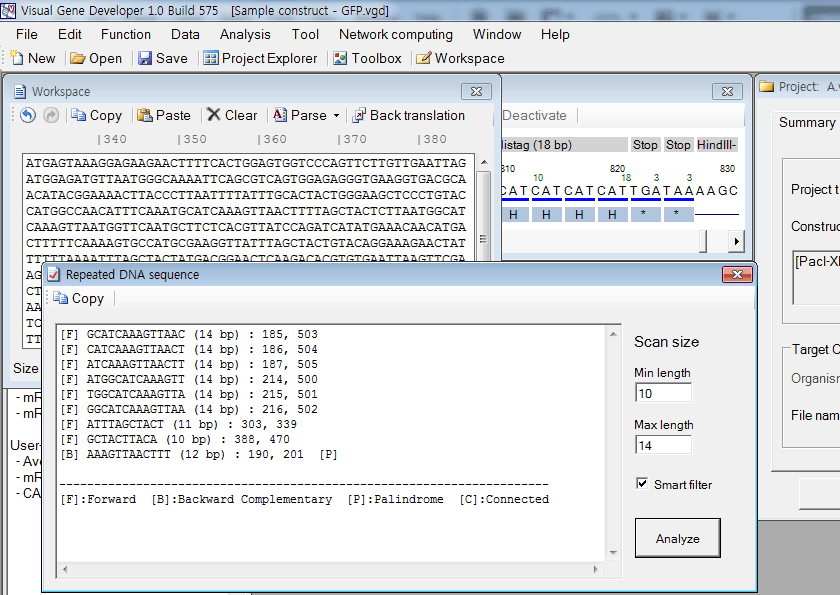
4. Meaning of letters
|
Letter |
Description |
| F | Forward direction search |
| B | Backward direction search |
| P | Palindromic sequence |
| C | There are duplicate repeated sequences that are connected to each other |
5. Scan size means the target length of repeated sequences to search. Recommended values are 10 for Minimum length and 10 for Maximum length, respectively.
6. If the 'Smart Filter' is checked on, duplicate repeated sequences will be removed from the list.
Silent removal of multiple sequences
* Silent removal means removal of certain target DNA sequence from a source sequence by replacing certain codons with synonymous codons while maintaining the expression of the same amino acids.
1. First input sequence to the textbox in the 'Workspace' window.
2. Click on the 'Silent removal of sequences' in the 'Function' menu.
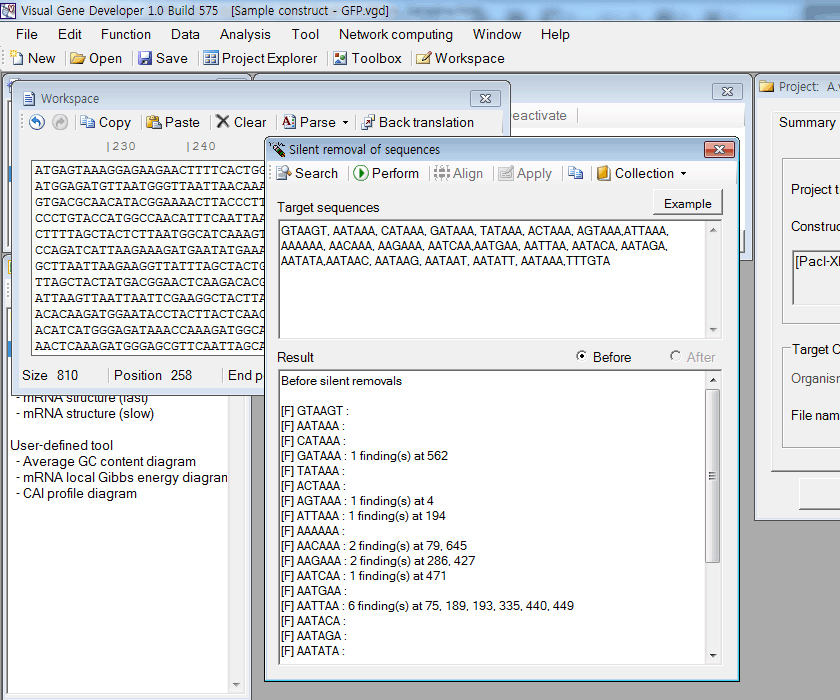
3. Put target sequences in the 'Target sequences' box. Each sequence can be separated by comma (,), Tab, or carriage return key (Enter key)
4. Firstly, click on the 'Search' button. You can see search result in the 'Result' box.
5. Secondly, click on the 'Perform' button. You can see the search result after silent removals in the 'Result' box
6. Thirdly, click on the 'Apply' button to overwrite a source sequence in the 'Workspace' window by a modified sequence
7. Optionally, you can compare the modified sequence with the original sequence. Just click on the 'Align' button.
Degenerate sequence
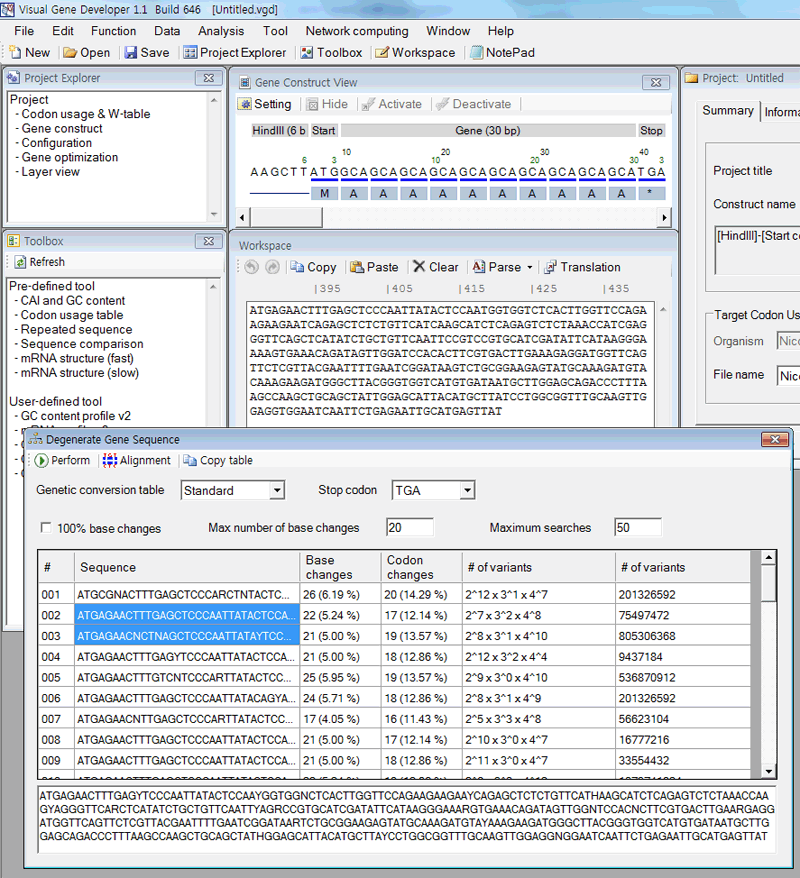
1. First input sequence to the textbox in the 'Workspace' window.
2. Click on the 'Degenerate gene sequence' in the 'Function' menu.
3. Click on the 'Perform' button in the 'Degenerate Gene Sequence' window.
Codon bias diagram of multiple genes
* Imitation of the job done by Plotkin and Kudla (Nature, 2011, p32-42)
1. We assume that there are multiple gene constructs in the 'Gene Optimization' window
Alternatively, a user can import sequences of multiple genes.
2. Choose a gene construct clicking on a cell in the 'Gene Optimization' window.
3. Click on 'Codon bias diagram' in the 'Toolbox' window
4. A pop-up window will be shown to ask the target gene component. Input a number!
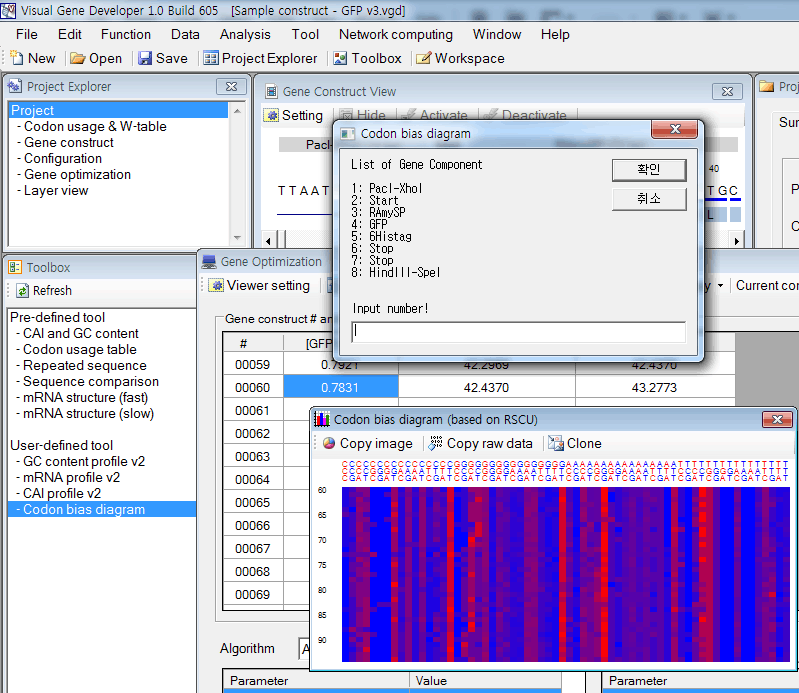
5. Then, you can see a new window titled 'Codon bias diagram (based on RSCU)'
* The function was developed using VBScript and its source code was revealed (Check Source).
Check Module Development section
Profiling functions: CAI, mRNA binding energy, tAI, and GC content
1. First of all, paste test sequence to a text box in the 'Workspace' window
2. Click on 'GC content profile v2', 'mRNA profile v2', 'tAI profile v1', or 'CAI profile v2' in the 'Toolbox' window
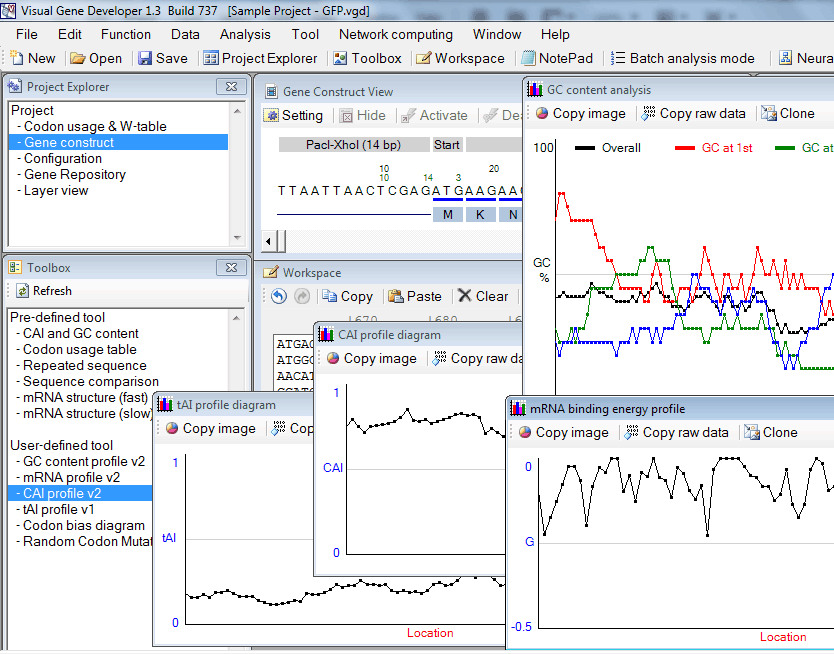
* Click on the 'Copy image' button to export image data to the clipboard
* Click on the 'Copy raw data' button to export tabulated text raw data to the clipboard
* Click on the 'Clone' button to generate a clone to keep the current result.
* The functions were developed using VBScript and its source code was revealed. Check 'Module Editor' or 'Module Library' window
For the profiling functions, a moving window method was equally adopted as shown in the following animation.
Ex). mRNA profiling function
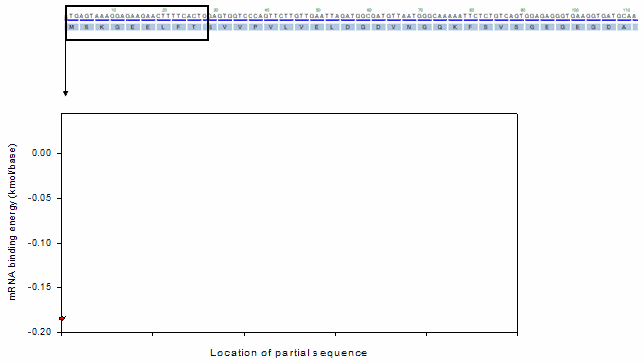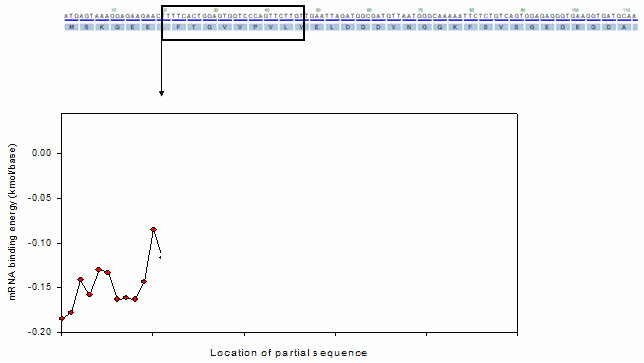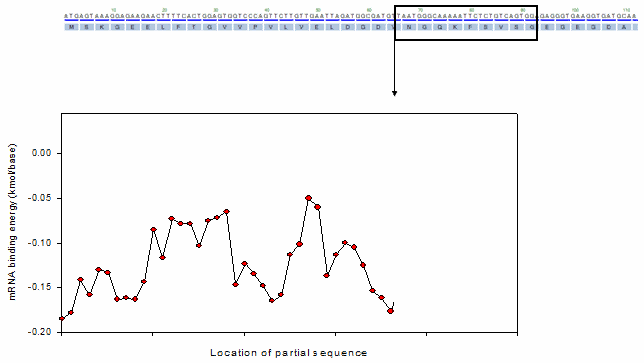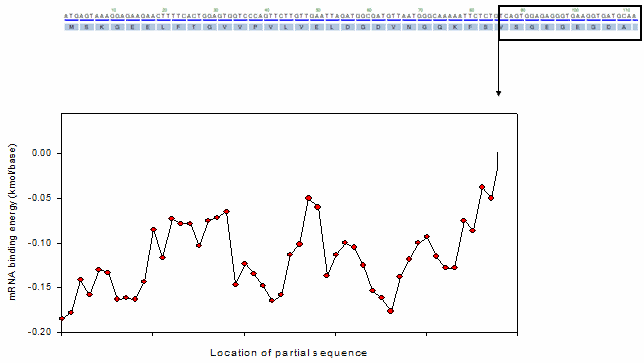
o Toolbox
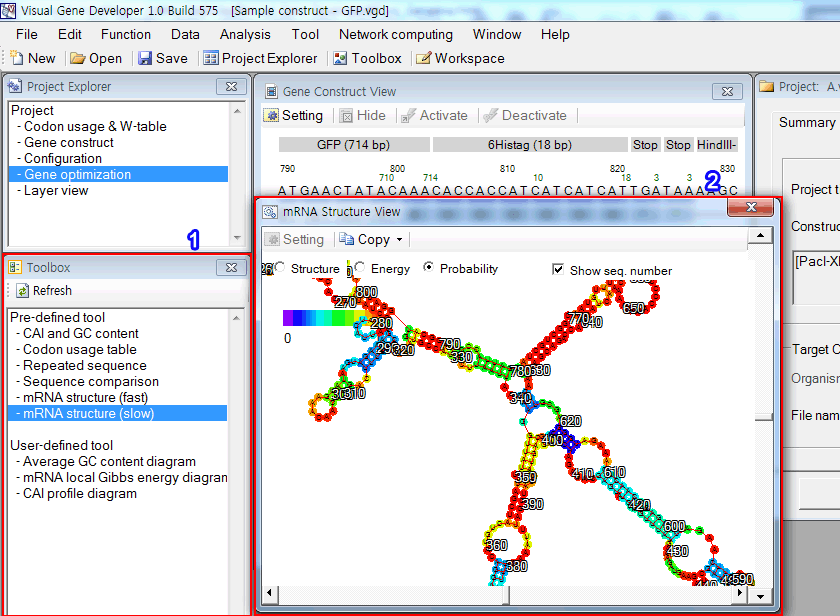
The 'Toolbox' window includes both predefined and user-defined functions. Just double click on an item in the list box.
For example,
1. Click on the 'mRNA structure (slow)'
2. You can see the predicted mRNA secondary structure in the new window
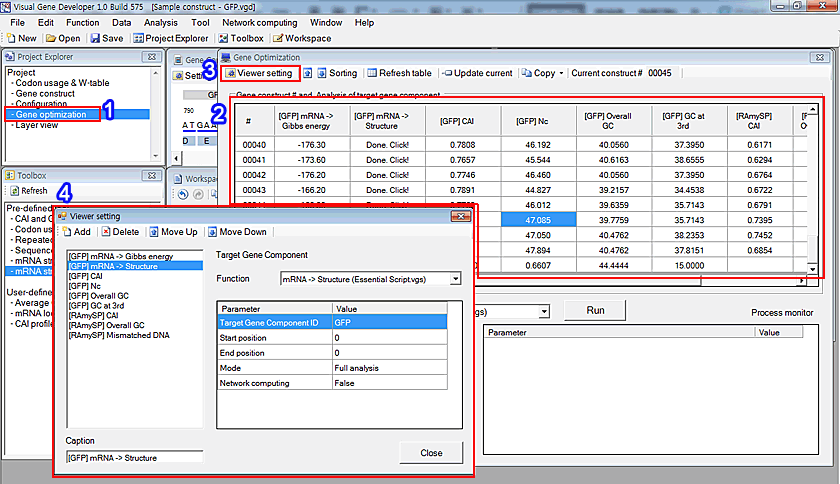
o Gene construct list
During the gene optimization process, all analysis results can be reviewed in the 'Gene optimization' window. A user can edit visible functions in the table in the 'Viewer setting' window.
1. Click on the 'Gene optimization' in the 'Project Explorer' window
2. You can see the table in the 'Gene optimization' window
3. Click on the 'Viewer setting' button
4. Click on the 'Add' button in the 'Viewer setting' window to add new functions. The added items will be immediately shown in the table (see number 2)
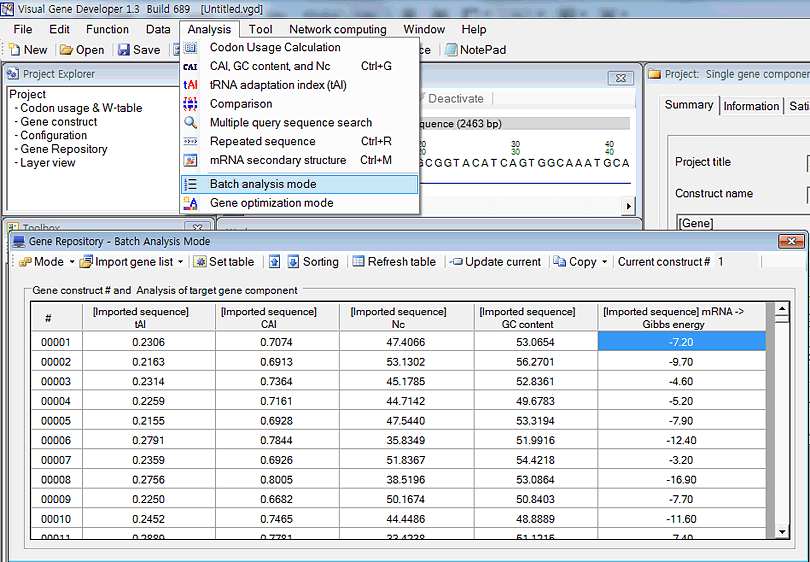
New update!
The 'Gene Optimization' window has been updated to 'Gene Repository' window
A user can choose convenient mode between 'Batch analysis mode' and 'Gene optimization mode'
o Module development and Class
All functions are implemented in a class that is a collection of modules. Programmatically, the class can be used when you develop modules.
For details, check the 'Class & module' section on the left menu.
Here is a sample we show.
1. Click on the 'Module library' in the 'Tool' menu
2. The 'Module Library' window will be shown. Choose 'GeneService_Calculate_CAI'
3. Click on the 'Edit Module' button
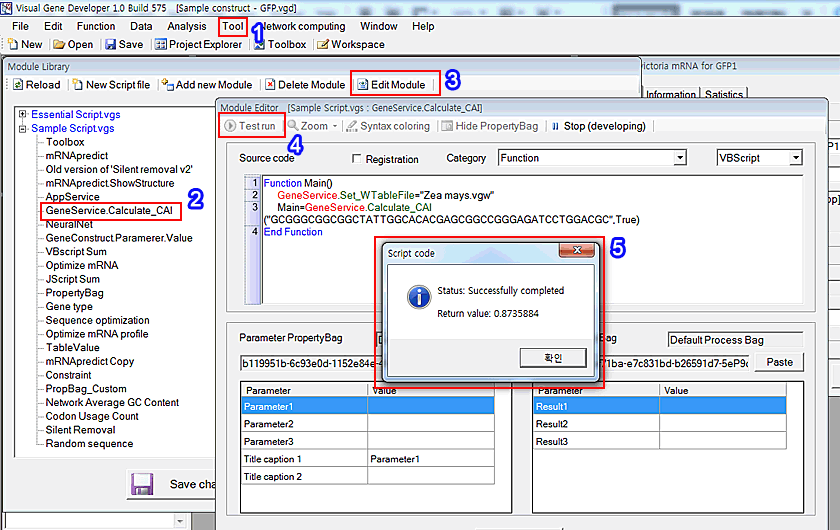
4. 'Module Editor' window will be pop-up. Click on the 'Test run' button.
Please check source code
VBScript
Function Main()
GeneService.Set_WTableFile="Zea mays.vgw"Main=GeneService.Calculate_CAI("GCGGGCGGCGGCTATTGGCACACGAGCGGCCGGGAGATCCTGGACGC",True)
End Function
JScript
function Main() {
GeneService.Set_WTableFile="Zea mays.vgw";
return GeneService.Calculate_CAI("GCGGGCGGCGGCTATTGGCACACGAGCGGCCGGGAGATCCTGGACGC","True");
}
'GeneService' is a class that includes lots of modules. 'Set-WTableFile' and 'Calculate_CAI' are modules of 'GeneService' class.
This simple example shows how to use the class and its functions (=modules) while you develop modules.
5. You can check the result in the 'Return message' window.
